Documentos de Académico
Documentos de Profesional
Documentos de Cultura
Advanced Enzymology Exercices
Cargado por
NaomiKerryaThompsonDerechos de autor
Formatos disponibles
Compartir este documento
Compartir o incrustar documentos
¿Le pareció útil este documento?
¿Este contenido es inapropiado?
Denunciar este documentoCopyright:
Formatos disponibles
Advanced Enzymology Exercices
Cargado por
NaomiKerryaThompsonCopyright:
Formatos disponibles
Exercices Advanced Enzymology
Y. Engelborghs
A. Diagnosis for inhibitors:
Four types of reversible inhibitors: competitive, uncompetitive, noncompetitive, mixed. To
identify the type of inhibitor we can construct a Lineweaver-Burk plot (1/v vs. 1/[S] ) for
with varying [I] and interpret the different meaning of the intercept and slope of the plot. For
a simple enzyme-substrate system the interpretation is:
Etot 1 K 1
= + M
v0 kcat kcat [ S ]
Intercept slope Slope/intercept
= 1/kcat KM/kcat KM
1. Competitive inhibition: equilibrium derivation:
Etot = [ E ] + [ ES ] + [ EI ]
Eactive = [ ES ]
[S ]
KM
v0 = vmax
[S ] [ I ]
1+ +
KM KI
Lineweaver Birk plot :
Etot 1 K M [ I ]K M
= ( + + 1)
v0 kcat [ S ] [ S ]K I
Identification of LB-parameters via Lineweaver-Birk plots for different values of [I]:
Advanced Enzymology Exercises - Y. Engelborghs - 1
Intercept Slope Slope/intercept
Competitive 1 KM [I ] [I ]
= Ct (1 + ) K M (1 + )
kcat kcat KI KI
2. Uncompetitive inhibition equilibrium derivation:
Etot = [ E ] + [ ES ] + [ ESI ]
Eactive = [ ES ]
[S ]
KM
v0 = vmax
[ S ] [ S ][ I ]
1+ +
KM KM KI
Lineweaver Birk plot :
Etot 1 KM [I ]
= ( + + 1)
v0 kcat [ S ] K I
Identification of LB-parameters:
Intercept Slope Slope/intercept
Uncompetitive 1 [I ] KM [I ]
(1 + ) = Ct K M /(1 + )
kcat KI kcat KI
Advanced Enzymology Exercises - Y. Engelborghs - 2
3. Non-competitive inhibition:
Etot = [ E ] + [ ES ] + [ EI ] + [ ESI ]
Eactive = [ ES ]
[S ]
KM
v0 = vmax
[ S ] [ I ] [ S ][ I ]
1+ + +
KM KI KM KI
Lineweaver Birk plot :
Etot 1 K M [ I ]K M [ I ]
= ( + + + 1)
v0 kcat [ S ] [ S ]K I K I
Identification of LB-parameters:
Intercept Slope Slope/intercept
noncompetitive 1
kcat
[I ]
(1 + )
KI
KM
kcat
[I ]
(1 + )
KI
K M = Ct
Advanced Enzymology Exercises - Y. Engelborghs - 3
4. Overall Diagnosis of Inhibitors:
Make primary Lineweaver-Birk plots of 1/v vs. 1/[A] for different values of [I]
Diagnosis: follow red diagonal �
Intercept Slope Slope/intercept
Competitive 1 KM [I ] [I ]
= Ct (1 + ) K M (1 + )
kcat kcat KI KI
Uncompetitive 1 [I ] KM [I ]
(1 + ) = Ct K M /(1 + )
v max KI kcat KI
noncompetitive 1
kcat
[I ]
(1 + )
KI
KM
kcat
[I ]
(1 + )
KI
K M = Ct
of primary Lineweaver-Birk plot
Logic:
competitive inhibition; Substrate can compete away I at high concentration,
therefore vmax = Ct
uncompetitive: I does not bind E therefore kcat/KM = Ct
(Mnemotechnic: uncompetitive I = I unactive at the unoccupied Enzyme)
Noncompetitive: I binds equally well to E as to ES therefore KM = Ct
Advanced Enzymology Exercises - Y. Engelborghs - 4
B. Diagnosis for Two substrate kinetics
With ternary complex :
- random sequence of binding;
- ordered mechanism
Without ternary complex:
- Ping pong mechanism : 2nd substrate binds after first
product formation
- Theorell Chance mechanism: a ternary complex is formed
but it is not populated because the 1st product formation is very fast.
1. Ternary complex formation with random order
Can be described via equilibrium assumption:
Etot = [ E ] + [ EA] + [ EB ] + [ EAB ]
Eactive = [ EAB ]
[ A][ B ]
K AKB f
v0 = vmax
[ A] [ B ] [ A][ B]
1+ + +
K A KB K AKB f
Lineweaver Birk plot :
Etot 1 K AKB f KB f K A f
= ( + + + 1)
v0 kcat [ A][ B ] [ B ] [ A]
f or 1 if A and B enforce each others binding then f<1, for a Briggs-Haldane
enzyme f may be >1.
Diagnosis: make primary Lineweaver-Birk plot with 1/v vs. 1/[A] with varying [B]:
Intercept Slope Slope/intercept
Random sequence 1 fK fK A K
(1 + B ) (1 + B ) K A if f = 1
kcat [ B] kcat [ B]
@Intercept
Advanced Enzymology Exercises - Y. Engelborghs - 5
Individual parameters can be obtained from secondary plots of intercepts or slopes vs.
1/[B]
Summary: information from secondary plots
Secondary plot intercept Slope Slope/intercept
Intercept vs 1/[B]-plot 1/kcat fKB/kcat fKB
Slope vs 1/[B]-plot fKA/kcat fKAKB/kcat KB
fKA f
KA
2. Ternary complex formation with ordered sequence (A obligatory
first)
Etot = [ E ] + [ EA] + [ EAB ]
Eactive = [ EAB ]
[ A][ B ]
K AKB
v0 = vmax
[ A] [ A][ B]
1+ +
K A K AKB
Lineweaver Birk plot :
Etot 1 K AKB KB
= ( + + 1)
v0 kcat [ A][ B] [ B]
Parameters of primary LB-plot:
Intercept Slope Slope/intercept
ordered 1 K K A KB K K
(1 + B ) ( ) K A B / 1 + B
v max [B] v max [ B ] [ B] [ B]
@Intercept =0
of primary Lineweaver-Birk plot
Advanced Enzymology Exercises - Y. Engelborghs - 6
3. For ping pong (enzyme substitution) mechanism
E + AX
EX + A
EX + B
E + BX
EX is a modified enzyme. The MM equation cannot be derived via equilibrium
assumptions, but has to be derived via transit-times (see notes). The result looks formally
very similar to the previous equation but there is no term (1) for E in the mass balance for
Etot
[ A][ B]
K AKB
v = vmax
[ A] [ B ] [ A][ B]
+ +
K A KB K AKB
Lineweaver Birk plot
Etot 1 KB K A
= ( + + 1)
v0 kcat [ B ] [ A]
Diagnostic: make Lineweaver-Birk plot with respect to [A]at varying
[B]. This leads to:
Intercept Slope Slope/intercept
Ping pong 1 K KA K
(1 + B ) = Ct K A /(1 + B )
kcat [ B] kcat [ B]
Parallel lines
Individual parameters can be obtained from secondary plots.
Advanced Enzymology Exercises - Y. Engelborghs - 7
KA
slope = independent of [ B] = primary LB plot : parallel lines
kcat
1 K 1
int ercept = (1 + B ) gives for [ B ]
kcat [ B] kcat
slope KA
= gives K A for [ B]
int ercept (1 + K B )
[ B]
Further proof for ping-pong: demonstrate existence of EX
4. Chance-Theorell: ternary complex is not populated kinetically
Cannot be distinguished from ordered reaction on the basis of initial rate studies
Advanced Enzymology Exercises - Y. Engelborghs - 8
5. Overall analysis two-substrate reactions:
Primary Lineweaver-Birkplot of 1/v vs. 1/[A]
Individual parameters can be obtained from secondary plots of intercept or slope vs 1/[B]
@ secondary plot of slope vs. 1/[B]
Intercept Slope Slope/intercept
Random sequence 1 fK fK A K
(1 + B ) (1 + B )
kcat [ B] kcat [ B]
@grows with
positive intercept
ordered 1 K K A KB K K
(1 + B ) ( ) K A B / 1 + B
kcat [ B] kcat [ B] [ B] [ B]
@grows with
intercept =0
Ping pong 1 KB KA K
(1 + ) = Ct K A /(1 + B )
kcat [ B] [ B]
kcat
@ remains ct
Theorell-Chance Similar to ordered
of primary Lineweaver-Birk plot
Advanced Enzymology Exercises - Y. Engelborghs - 9
C. Exercises
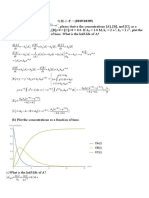
Exercise NR 1 acetylcholinesterase (Price & Dwek, p184)
The Effect of choline (C) on the reaction catalysed by acetylcholine-esterase was studied and
gave the following results: [AC] = acetylcholine concentration
[AC] mM [AC] mM [AC] mM [AC] mM [AC] mM
[C] mM 0.1 0.15 0.25 0.40 0.70
0 28.5@ 37.5 50.0 61.5 73.7
20 11.9 15.6 20.9 25.7 30.7
40 7.5 9.9 13.2 16.2 19.4
@ relative velocity (in arbitrary units)
Task: Analyse the data
Primary Lineweaver-Birk plots
acetylcholinesterase
0.14
0.12
0.10
0.08
1/v
0.06
0.04
0.02
0.00
0 2 4 6 8 10 12
1/[AC] 1/mM
C=0
C=20
C=40
Figure 1: Lineweaver-Birk plot for different [C]
Advanced Enzymology Exercises - Y. Engelborghs - 10
1) vmax is not constant but decreases when C increases = not competitive inhibition.
2) Intercepts and slopes increase when [C] increases !!!
3) Slope/intercept= constant = KMAC is independent on C: therefore : non competitive
inhibition
Intercept Slope Slope/intercept
C=0 0,98 x 10-2 2,58 x 10-3 0,263
C=20 2,36x 10-2 5,93 x 10-3 0,251
C=40 3,9x 10-2 9,35 x 10-3 0,24
Conclusion: KM = constant noncompetitive inhibition . Make secondary plots to get
parameters:
Exercise NR2 Fructose bisphosphate (FBP)
Fructose bisphosphatase (reaction catalysed?) is inhibited by AMP. The following data were
obtained from a study of the rat liver enzyme:
uM = micromolar
[FBP] uM [FBP] uM [FBP] uM [FBP] uM [FBP] uM
[AMP] uM 4 6 10 20 40
0 0.059@ 0.076 0.101 0.125 0.150
8 0.034 0.043 0.056 0.071 0.083
@ velocity in katal.kg-1
Analyse these data.
_________________________________
Exercise NR3 Nucleoside diphosphokinase
The enzyme nucleoside diphosphokinase will catalyse the following reaction:
GTP + dGDP GDP + dGTP in presence of Mg2+
In an experiment with an enzyme isolated from erythrocytes, the following result were
obtained:
[GTP] uM [GTP] uM [GTP] uM [GTP] uM
[dGDP]uM 22 30 50 200
20 0.095 0.112 0.141 0.196
25 0.102@ 0.120 0.155 0.223
40 0.112 0.136 0.180 0.284
100 0.125 0.156 0.218 0.385
Advanced Enzymology Exercises - Y. Engelborghs - 11
@ velocity in katal.kg-1
What is the likely mechanism for the enzyme? How could you check your conclusion?
________________________________________________________________________
Exercise NR4 Creatine Kinase
A study was made of the reaction catalysed by creatine kinase.
Creatine + ATP phosphocreatine + ADP (in the presence of Mg2+ )
The following data were obtained:
[ATP] mM [ATP] mM [ATP] mM [ATP] mM
0.46 0.62 1.23 3.68
[creatine] mM
6 0.38@ 0.46 0.66 0.97
10 0.55 0.68 0.95 1.31
20 0.85 1.00 1.34 1.80
40 1.18 1.38 1.72 2.29
@ velocity in katal.kg-1
Evaluate the kinetic parameters for this reaction. From other data it appears that the
reaction proceeds via a random order ternary complex mechanism. What can you deduce
about the binding of substrates to the enzyme?
______________________________________________________________________
Exercise Nr 5 Alcohol deydrogenase
The following data were obtained in a study of the reaction catalysed by yeast alcohol
dehydrogenase:
Ethanol + NAD+ acetaldehyde + NADH
[NAD+] mM [NAD+] mM [NAD+] mM [NAD+] mM
[Ethanol] mM 0.05 0.10 0.25 1.0
10 0.30@ 0.51 0.89 1.43
20 0.44 0.75 1.32 2.11
40 0.57 1.00 1.72 2.76
200 0.76 1.31 2.29 3.67
@ velocity in katal.kg-1
Determine the mechanism and the kinetic parameters of this reaction.
Advanced Enzymology Exercises - Y. Engelborghs - 12
Exercise NR6
The effect of the products (pyrvate) on the reaction catalysed by rabbit muscle lactate
dehydrogenase was studied, with the results in the following tables.
Reaction: NAD+ + lactate NADH + pyruvate
Experiment 1: Following data were obtained at fixed concentration of [NAD+] = 1,5
mM
[Lact] mM [Lact] mM [Lact] mM [Lact] mM
[Pyruvate] uM 1.5 2.0 3.0 10.0
0 1.88@ 2.36 3.10 5.81
40 1.05 1.34 1.88 4.19
80 0.73 0.94 1.34 3.27
@ velocity in katal.kg-1
Experiment 2: Following data were obtained at fixed concentration of Lactate [Lact] = 15
mM
[NAD+] mM [NAD+] mM [NAD+] mM [NAD+] mM
[Pyruvate] uM 0.5 0.7 1.0 2.0
0 3.33@ 3.91 4.50 5.43
30 2.65 3.13 3.60 4.33
60 1.97 2.30 2.66 3.21
@ velocity in katal.kg-1
What is the interpretation of these data?
Advanced Enzymology Exercises - Y. Engelborghs - 13
Exercise NR 7
Chymotrypsin catalyses the hydrolysis of p-Nitrophenolacetate (PNPA) to yield
p-nitrophenol and acetate. The production of p-nitrophenol (PNP) was followed as a
function of time with the following results:
experiment 1: [chymotrypsin]= 23 uM; [PNPA] = 500 uM
t(s) 0 15 30 60 90 120 150 180
[PNP]uM 0 18.3 22.2 29.1 35.7 41.7 48.0 54.3
Experiment 2: chymotrypsin]= 12 uM; [PNPA] = 500 uM
t(s) 0 15 30 60 90 120 150 180
[PNP]uM 0 10.3 12.6 16.2 19.7 23.3 26.8 30.4
What do you deduce from these data?
Advanced Enzymology Exercises - Y. Engelborghs - 14
También podría gustarte
- How To Believe God For A House Study NotesDocumento47 páginasHow To Believe God For A House Study NotesJamie Mary Peter95% (61)
- Multiplication 5 Minute Frenzy (A)Documento1 páginaMultiplication 5 Minute Frenzy (A)NaomiKerryaThompsonAún no hay calificaciones
- U2 c1 AnswersDocumento8 páginasU2 c1 AnswersMaiah Phylicia Latoya50% (2)
- Solution Manual for an Introduction to Equilibrium ThermodynamicsDe EverandSolution Manual for an Introduction to Equilibrium ThermodynamicsAún no hay calificaciones
- MCAT AAMC Content Outline - Science PDFDocumento165 páginasMCAT AAMC Content Outline - Science PDFadfhAún no hay calificaciones
- Toxicokinetics and Saturation KineticsDocumento51 páginasToxicokinetics and Saturation KineticsSyama J.S75% (4)
- Amylase Experiment Lab ReportDocumento7 páginasAmylase Experiment Lab ReportCHLOE IANNAH CALVADORESAún no hay calificaciones
- CatalysisDocumento82 páginasCatalysisDewan1100% (1)
- Nonlinear Pharmacokinetics 1Documento11 páginasNonlinear Pharmacokinetics 1Mubashar ShahidAún no hay calificaciones
- DAVAO BiochemDocumento5 páginasDAVAO BiochemVince Cabahug100% (1)
- GCSE Maths - EquationsDocumento50 páginasGCSE Maths - EquationsPrinceShalomAún no hay calificaciones
- Matlab Simulink DC MotorDocumento12 páginasMatlab Simulink DC Motorkillua142100% (4)
- Enzyme Kinetics of Alpha-AmylaseDocumento8 páginasEnzyme Kinetics of Alpha-AmylaseJasmin CeciliaAún no hay calificaciones
- Enzyme InhibitionDocumento20 páginasEnzyme InhibitionAnkita guptaAún no hay calificaciones
- 6 Behavior of Proteins Enzymes PDFDocumento35 páginas6 Behavior of Proteins Enzymes PDFKelly SisonAún no hay calificaciones
- 21.8 (A) Lindermann-Hinshelwood Mechanism: (Slow, RDS) + +Documento18 páginas21.8 (A) Lindermann-Hinshelwood Mechanism: (Slow, RDS) + +cmc107Aún no hay calificaciones
- Michaelis-Menten Model Michaelis-Menten Model (Cont'd) : - For An Enzyme-Catalyzed ReactionDocumento2 páginasMichaelis-Menten Model Michaelis-Menten Model (Cont'd) : - For An Enzyme-Catalyzed ReactionAciel CruzAún no hay calificaciones
- Rumus Dan Persamaan Yang Dapat Digunakan (Soal Farmakokinetika)Documento1 páginaRumus Dan Persamaan Yang Dapat Digunakan (Soal Farmakokinetika)asriAún no hay calificaciones
- Kinetics Formula SheetDocumento1 páginaKinetics Formula SheetdfasdfAún no hay calificaciones
- 5.enzyme Inhibition-TransDocumento24 páginas5.enzyme Inhibition-TransSuneo Yang ComelAún no hay calificaciones
- Chapter 14. Enzyme KineticsDocumento44 páginasChapter 14. Enzyme KineticsKalai VaniAún no hay calificaciones
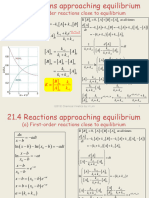
- (A) First-Order Reactions Close To Equilibrium: Da Ka K B DT K Ke A A K KDocumento18 páginas(A) First-Order Reactions Close To Equilibrium: Da Ka K B DT K Ke A A K Kcmc107Aún no hay calificaciones
- 1 Ti T 0 de (T) DTDocumento1 página1 Ti T 0 de (T) DTUsha RAún no hay calificaciones
- Enzyme KineticsDocumento4 páginasEnzyme KineticsvnAún no hay calificaciones
- Lecture3 (Adsorption-Part2) 2022Documento8 páginasLecture3 (Adsorption-Part2) 2022최종윤Aún no hay calificaciones
- (Pre-Equilibrium) : AB K B A KDocumento9 páginas(Pre-Equilibrium) : AB K B A KHarsh KajiAún no hay calificaciones
- 20.320 Exam 2 Review Problem Solutions April 20, 2006 1. No InhibitionDocumento3 páginas20.320 Exam 2 Review Problem Solutions April 20, 2006 1. No InhibitionMovie Scene BankAún no hay calificaciones
- Solving Real Business Cycle Models by Solving Systems of First Order ConditionsDocumento18 páginasSolving Real Business Cycle Models by Solving Systems of First Order ConditionsBrunéAún no hay calificaciones
- Lecture 1Documento8 páginasLecture 1sabbithiAún no hay calificaciones
- Macro Theory110 Assignment 5Documento8 páginasMacro Theory110 Assignment 5Greco S50Aún no hay calificaciones
- ParallRxn PDFDocumento1 páginaParallRxn PDFWizra QamarAún no hay calificaciones
- ParallRxn PDFDocumento1 páginaParallRxn PDFAbhinav RajeshAún no hay calificaciones
- ParallRxn PDFDocumento1 páginaParallRxn PDFAbhinav RajeshAún no hay calificaciones
- Assignment - 2 Sol PDFDocumento7 páginasAssignment - 2 Sol PDFGreco S50Aún no hay calificaciones
- 108 Quiz1Documento1 página108 Quiz1沈智恩Aún no hay calificaciones
- Enzyme InhibitionDocumento13 páginasEnzyme InhibitionMheira Villahermosa100% (1)
- Mathematical Relations PDFDocumento229 páginasMathematical Relations PDFmars100% (1)
- Nuggets: Evaluating Rate Laws: 3. Integrated Rate Laws, 4. Half-Lives, 5. Rate Versus ConcentrationDocumento12 páginasNuggets: Evaluating Rate Laws: 3. Integrated Rate Laws, 4. Half-Lives, 5. Rate Versus ConcentrationRosario QFAún no hay calificaciones
- DCS Proj1 2024Documento2 páginasDCS Proj1 2024blackman510101Aún no hay calificaciones
- Enzyme KineticsDocumento65 páginasEnzyme KineticsSulabh JainAún no hay calificaciones
- Ligand Binding: A. Binding To A Single SiteDocumento8 páginasLigand Binding: A. Binding To A Single SiteRahul shyamAún no hay calificaciones
- Tutorial Sol CH 8Documento6 páginasTutorial Sol CH 8Abraham wisdomAún no hay calificaciones
- 5.62 Physical Chemistry Ii: Mit OpencoursewareDocumento8 páginas5.62 Physical Chemistry Ii: Mit OpencoursewareavdvAún no hay calificaciones
- 10 Enzymatic ReactionDocumento30 páginas10 Enzymatic ReactionWidya Nur RamadhaniAún no hay calificaciones
- 2015-Im-25 Ic CepDocumento25 páginas2015-Im-25 Ic Cepzaib hassanAún no hay calificaciones
- Assignment #2 - Self-Learning SlidesDocumento4 páginasAssignment #2 - Self-Learning Slidesbebble boppleAún no hay calificaciones
- Assignment - 3 - Ans (Updated)Documento6 páginasAssignment - 3 - Ans (Updated)Greco S50Aún no hay calificaciones
- 1 Lecture Notes: The Solow ModelDocumento5 páginas1 Lecture Notes: The Solow Modelmrs dosadoAún no hay calificaciones
- Assignment: (Document Subtitle)Documento11 páginasAssignment: (Document Subtitle)aqeelAún no hay calificaciones
- Generalised Minimum Variance ControllerDocumento32 páginasGeneralised Minimum Variance Controllerenzo velasquezAún no hay calificaciones
- CPII Enzymatic KineticsDocumento13 páginasCPII Enzymatic KineticsValentina CretuAún no hay calificaciones
- Che 407 TolDocumento4 páginasChe 407 Toldavidolalere7Aún no hay calificaciones
- SolutionDocumento3 páginasSolutionJocelyn garcia gonzalezAún no hay calificaciones
- Real Business Cycles: Jes Us Fern Andez-Villaverde University of PennsylvaniaDocumento22 páginasReal Business Cycles: Jes Us Fern Andez-Villaverde University of PennsylvaniawilianssterAún no hay calificaciones
- Laplace AnaliticaDocumento2 páginasLaplace AnaliticaValaur Ekbalam M BAún no hay calificaciones
- 56 - Comments On "Antiwindup Strategy For PI-Type Speed Controller"Documento4 páginas56 - Comments On "Antiwindup Strategy For PI-Type Speed Controller"Infy WangAún no hay calificaciones
- Velocity Lab ReportDocumento15 páginasVelocity Lab ReportchrisAún no hay calificaciones
- Solution Manual For Physical Chemistry Thermodynamics Statistical Mechanics and Kinetics 1St Edition Andrew Cooksy 0321814150 Full Chapter PDFDocumento36 páginasSolution Manual For Physical Chemistry Thermodynamics Statistical Mechanics and Kinetics 1St Edition Andrew Cooksy 0321814150 Full Chapter PDFwilliam.shifflett812100% (14)
- MEP Is LM EquationsDocumento12 páginasMEP Is LM EquationsShreyas SatardekarAún no hay calificaciones
- Macroeconomics 1 PS3 Solution PDFDocumento10 páginasMacroeconomics 1 PS3 Solution PDFTaib MuffakAún no hay calificaciones
- DiscreteTime MPC (2021)Documento20 páginasDiscreteTime MPC (2021)Kamgang BlaiseAún no hay calificaciones
- Position LabDocumento15 páginasPosition LabchrisAún no hay calificaciones
- Assignment 5 VFI PDFDocumento2 páginasAssignment 5 VFI PDFGreco S50Aún no hay calificaciones
- NnjjbyvpDocumento7 páginasNnjjbyvpamin badrulAún no hay calificaciones
- 2 B - Ecuacion de Euler Y Condicion de TransversalidadDocumento4 páginas2 B - Ecuacion de Euler Y Condicion de TransversalidadGonzalo del FierroAún no hay calificaciones
- Dynare RBCDocumento21 páginasDynare RBCc9bj9bvr5dAún no hay calificaciones
- Standard Five Pupil Aces CSEC Math - Grade ADocumento2 páginasStandard Five Pupil Aces CSEC Math - Grade ANaomiKerryaThompsonAún no hay calificaciones
- SCD Application ChecklistDocumento1 páginaSCD Application ChecklistNaomiKerryaThompsonAún no hay calificaciones
- Applied Maths Model Questions Set 1Documento24 páginasApplied Maths Model Questions Set 1Courtney Taylor0% (1)
- J. Antimicrob. Chemother. 2001 Andrews 5 16Documento0 páginasJ. Antimicrob. Chemother. 2001 Andrews 5 16fatih28kaiserAún no hay calificaciones
- Maths Review TestDocumento5 páginasMaths Review TestNaomiKerryaThompsonAún no hay calificaciones
- Multiplication 5 Minute Frenzy (B)Documento1 páginaMultiplication 5 Minute Frenzy (B)NaomiKerryaThompsonAún no hay calificaciones
- Application Form PSTA PostsDocumento4 páginasApplication Form PSTA PostsNaomiKerryaThompsonAún no hay calificaciones
- Maths Review TestDocumento5 páginasMaths Review TestNaomiKerryaThompsonAún no hay calificaciones
- Times Table 1-4Documento1 páginaTimes Table 1-4NaomiKerryaThompsonAún no hay calificaciones
- Application Form PSTA PostsDocumento4 páginasApplication Form PSTA PostsNaomiKerryaThompsonAún no hay calificaciones
- Multiplication Pop QuizDocumento2 páginasMultiplication Pop QuizNaomiKerryaThompsonAún no hay calificaciones
- Protein and Amino AcidsDocumento5 páginasProtein and Amino AcidsMia SolimanAún no hay calificaciones
- Strand 8: Number TheoryDocumento21 páginasStrand 8: Number TheoryNaomiKerryaThompsonAún no hay calificaciones
- English Review TestDocumento5 páginasEnglish Review TestNaomiKerryaThompsonAún no hay calificaciones
- Application Form PSTA PostsDocumento4 páginasApplication Form PSTA PostsNaomiKerryaThompsonAún no hay calificaciones
- Student Text: UNIT 7 Number System and Bases AnswersDocumento5 páginasStudent Text: UNIT 7 Number System and Bases Answersapi-195130729Aún no hay calificaciones
- Student Text: UNIT 7 Number System and Bases AnswersDocumento5 páginasStudent Text: UNIT 7 Number System and Bases Answersapi-195130729Aún no hay calificaciones
- Sea Guidelines PDFDocumento28 páginasSea Guidelines PDFNaomiKerryaThompsonAún no hay calificaciones
- Sea Guidelines PDFDocumento28 páginasSea Guidelines PDFNaomiKerryaThompsonAún no hay calificaciones
- Squares, Cubes, Exponents, Place Value WorksheetDocumento5 páginasSquares, Cubes, Exponents, Place Value WorksheetNaomiKerryaThompsonAún no hay calificaciones
- UNIT 7 Number System and Bases: Introduction To SIMDocumento2 páginasUNIT 7 Number System and Bases: Introduction To SIMNaomiKerryaThompsonAún no hay calificaciones
- Times Table Quiz 1-4Documento1 páginaTimes Table Quiz 1-4NaomiKerryaThompsonAún no hay calificaciones
- Times Table 5-8Documento1 páginaTimes Table 5-8NaomiKerryaThompsonAún no hay calificaciones
- Division Tables Table 1-4Documento1 páginaDivision Tables Table 1-4NaomiKerryaThompsonAún no hay calificaciones
- Multiplication & Division With NegativesDocumento2 páginasMultiplication & Division With NegativesNaomiKerryaThompsonAún no hay calificaciones
- Protein and Amino AcidsDocumento5 páginasProtein and Amino AcidsMia SolimanAún no hay calificaciones
- 3 Enzymes - MCQ QPDocumento4 páginas3 Enzymes - MCQ QPVivehaAún no hay calificaciones
- IVMS Pharmacology Flash FactsDocumento8250 páginasIVMS Pharmacology Flash FactsMarc Imhotep Cray, M.D.Aún no hay calificaciones
- Chapter Two Enzyme Kinetics: Tesfayesus ZinareDocumento54 páginasChapter Two Enzyme Kinetics: Tesfayesus ZinarehabteAún no hay calificaciones
- L08 - Enzyme Kinetics IDocumento34 páginasL08 - Enzyme Kinetics ILeroy ChengAún no hay calificaciones
- Cecil Instruments Ce 7400 BrochureDocumento12 páginasCecil Instruments Ce 7400 BrochureDaniela Tanase100% (2)
- Teknik ReaksiDocumento21 páginasTeknik ReaksiAhmadi FachryAún no hay calificaciones
- ALL Chemistry CarbohydratesDocumento70 páginasALL Chemistry CarbohydratesPiyush Bhandari100% (1)
- Ch06 LO Su18Documento10 páginasCh06 LO Su18Mariam EidAún no hay calificaciones
- Enzyme Assays and KineticsDocumento12 páginasEnzyme Assays and KineticsKNTAún no hay calificaciones
- From By-Product To Valuable Components: Efficient Enzymatic Conversion of Lactose in Whey Using Galactosidase From Streptococcus ThermophilusDocumento9 páginasFrom By-Product To Valuable Components: Efficient Enzymatic Conversion of Lactose in Whey Using Galactosidase From Streptococcus ThermophilusAntonela PortaAún no hay calificaciones
- ToxicokineticsDocumento17 páginasToxicokineticsAshwin Ben JamesAún no hay calificaciones
- Analysis of Film and Pore Diffusion Effects On Kinetics of Immobilized Enzyme ReactionsDocumento7 páginasAnalysis of Film and Pore Diffusion Effects On Kinetics of Immobilized Enzyme ReactionsThirunavukkarasu ArunachalamAún no hay calificaciones
- Solution Manual For Biochemistry A Short Course 3rd by TymoczkoDocumento7 páginasSolution Manual For Biochemistry A Short Course 3rd by TymoczkoAmy Carter100% (37)
- Lecture 3 - Monod KineticsDocumento19 páginasLecture 3 - Monod KineticsZeny Naranjo0% (1)
- 9 Enzyme KineticsDocumento16 páginas9 Enzyme Kineticsaerum deijiAún no hay calificaciones
- Enzymes: OutlineDocumento10 páginasEnzymes: OutlineManila MedAún no hay calificaciones
- The Effects of Temperature, PH and Metal Ions On Alpha Amylase Activity of The Brine Shrimp Artemia SalinaDocumento11 páginasThe Effects of Temperature, PH and Metal Ions On Alpha Amylase Activity of The Brine Shrimp Artemia SalinaMarco Ramos0% (1)
- Biochemistry Lab ReportDocumento8 páginasBiochemistry Lab ReportEdward50% (2)
- Mechaelis-Menten KineticsDocumento7 páginasMechaelis-Menten KineticsfayeAún no hay calificaciones
- Chapter 23Documento53 páginasChapter 23haririAún no hay calificaciones
- BABS3031/2631 Equation SheetDocumento5 páginasBABS3031/2631 Equation SheetFlame ShineAún no hay calificaciones
- Colorimetric Detection of Hydrogen Peroxide and Glucose by Exploiting The Peroxidase-Like Activity of PapainDocumento5 páginasColorimetric Detection of Hydrogen Peroxide and Glucose by Exploiting The Peroxidase-Like Activity of PapainPhat HuynhAún no hay calificaciones
- Quiz On Enzyme KineticsDocumento3 páginasQuiz On Enzyme KineticsHens Christian FuentesAún no hay calificaciones